备注
Go to the end 下载完整的示例代码。或者通过浏览器中的MysterLite或Binder运行此示例
梯度增强回归#
此示例演示梯度提升从弱预测模型集合生成预测模型。梯度提升可用于回归和分类问题。在这里,我们将训练一个模型来处理糖尿病回归任务。我们将从以下方面获得结果: GradientBoostingRegressor 具有最小平方损失和深度为4的500棵回归树。
注:对于更大的数据集(n_samples >= 10000),请参阅 HistGradientBoostingRegressor .看到 梯度增强树的梯度中的功能 举个例子,展示了 HistGradientBoostingRegressor .
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
from sklearn import datasets, ensemble
from sklearn.inspection import permutation_importance
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import train_test_split
from sklearn.utils.fixes import parse_version
加载数据#
首先我们需要加载数据。
diabetes = datasets.load_diabetes()
X, y = diabetes.data, diabetes.target
数据预处理#
接下来,我们将拆分数据集,将90%用于训练,其余部分用于测试。我们还将设置回归模型参数。您可以使用这些参数来查看结果如何变化。
n_estimators :将执行的升压级的数量。稍后,我们将绘制与提升迭代的偏差。
max_depth :限制树中的节点数量。最佳值取决于输入变量的相互作用。
min_samples_split :拆分内部节点所需的最小样本数。
learning_rate :每棵树的贡献会减少多少。
loss : loss function to optimize. The least squares function is used in this case however, there are many other options (see GradientBoostingRegressor ).
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.1, random_state=13
)
params = {
"n_estimators": 500,
"max_depth": 4,
"min_samples_split": 5,
"learning_rate": 0.01,
"loss": "squared_error",
}
匹配回归模型#
现在我们将启动梯度增强回归器,并将其与我们的训练数据进行匹配。让我们再看看测试数据的均方误差。
reg = ensemble.GradientBoostingRegressor(**params)
reg.fit(X_train, y_train)
mse = mean_squared_error(y_test, reg.predict(X_test))
print("The mean squared error (MSE) on test set: {:.4f}".format(mse))
The mean squared error (MSE) on test set: 3010.2061
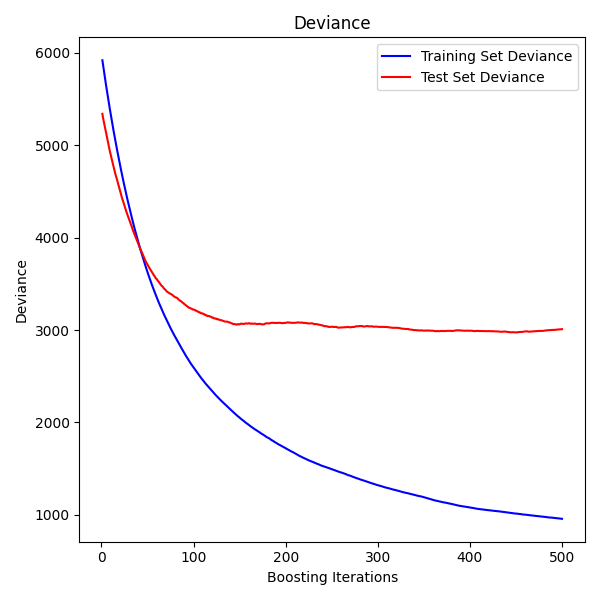
情节训练偏差#
最后,我们将可视化结果。为此,我们将首先计算测试集偏差,然后根据增强迭代绘制它。
test_score = np.zeros((params["n_estimators"],), dtype=np.float64)
for i, y_pred in enumerate(reg.staged_predict(X_test)):
test_score[i] = mean_squared_error(y_test, y_pred)
fig = plt.figure(figsize=(6, 6))
plt.subplot(1, 1, 1)
plt.title("Deviance")
plt.plot(
np.arange(params["n_estimators"]) + 1,
reg.train_score_,
"b-",
label="Training Set Deviance",
)
plt.plot(
np.arange(params["n_estimators"]) + 1, test_score, "r-", label="Test Set Deviance"
)
plt.legend(loc="upper right")
plt.xlabel("Boosting Iterations")
plt.ylabel("Deviance")
fig.tight_layout()
plt.show()
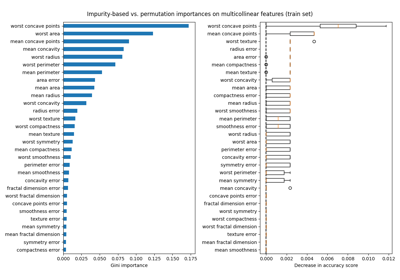
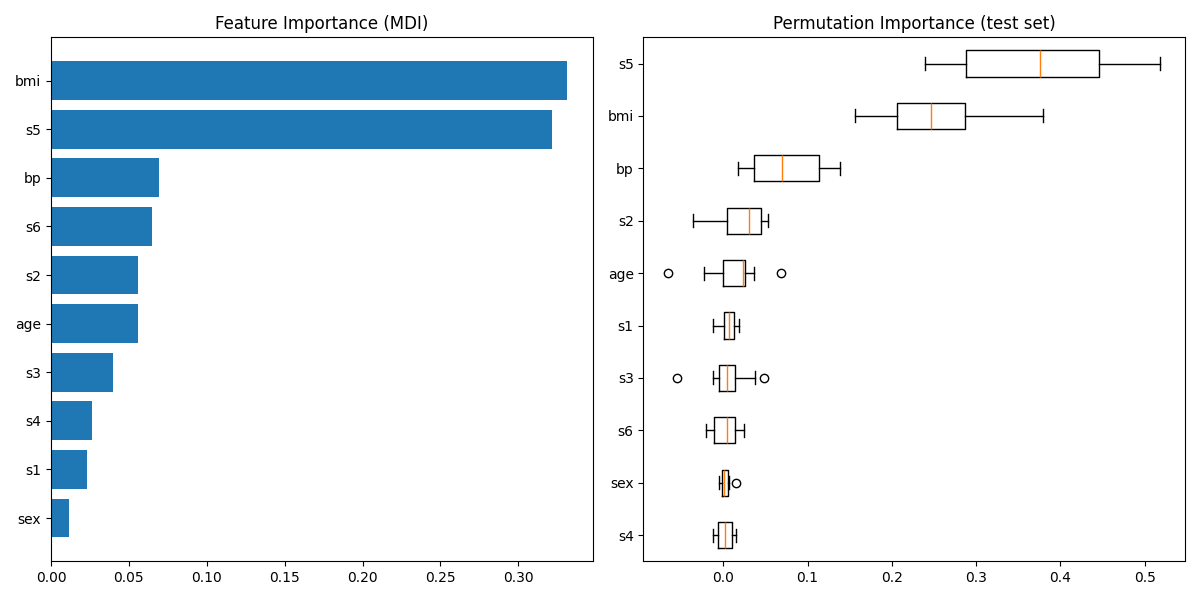
情节特征重要性#
警告
Careful, impurity-based feature importances can be misleading for
high cardinality features (many unique values). As an alternative,
the permutation importances of reg can be computed on a
held out test set. See 排列特征重要性 for more details.
对于本例,基于杂质和排列的方法识别相同的2个强预测特征,但顺序不同。第三个最具预测性的特征“BP”对于这2种方法也是相同的。其余特征的预测性较差,排列图的误差线显示它们与0重叠。
feature_importance = reg.feature_importances_
sorted_idx = np.argsort(feature_importance)
pos = np.arange(sorted_idx.shape[0]) + 0.5
fig = plt.figure(figsize=(12, 6))
plt.subplot(1, 2, 1)
plt.barh(pos, feature_importance[sorted_idx], align="center")
plt.yticks(pos, np.array(diabetes.feature_names)[sorted_idx])
plt.title("Feature Importance (MDI)")
result = permutation_importance(
reg, X_test, y_test, n_repeats=10, random_state=42, n_jobs=2
)
sorted_idx = result.importances_mean.argsort()
plt.subplot(1, 2, 2)
# `labels` argument in boxplot is deprecated in matplotlib 3.9 and has been
# renamed to `tick_labels`. The following code handles this, but as a
# scikit-learn user you probably can write simpler code by using `labels=...`
# (matplotlib < 3.9) or `tick_labels=...` (matplotlib >= 3.9).
tick_labels_parameter_name = (
"tick_labels"
if parse_version(matplotlib.__version__) >= parse_version("3.9")
else "labels"
)
tick_labels_dict = {
tick_labels_parameter_name: np.array(diabetes.feature_names)[sorted_idx]
}
plt.boxplot(result.importances[sorted_idx].T, vert=False, **tick_labels_dict)
plt.title("Permutation Importance (test set)")
fig.tight_layout()
plt.show()
Total running time of the script: (0分1.202秒)
相关实例
Gallery generated by Sphinx-Gallery <https://sphinx-gallery.github.io> _