备注
Go to the end 下载完整的示例代码。或者通过浏览器中的MysterLite或Binder运行此示例
随机投影嵌入的Johnson-Lindenstrauss界#
的 Johnson-Lindenstrauss lemma 指出任何高维数据集都可以随机投影到低维欧氏空间中,同时控制成对距离的失真。
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
import sys
from time import time
import matplotlib.pyplot as plt
import numpy as np
from sklearn.datasets import fetch_20newsgroups_vectorized, load_digits
from sklearn.metrics.pairwise import euclidean_distances
from sklearn.random_projection import (
SparseRandomProjection,
johnson_lindenstrauss_min_dim,
)
理论性能边界#
随机投影引入的失真 p 事实证明, p 正在以良好的概率定义eps嵌入,定义如下:
哪里 u 和 v 是从形状数据集中获取的任何行 (n_samples, n_features) 和 p 是一个随机高斯分布的投影 N(0, 1) 形状矩阵 (n_components, n_features) (or稀疏Achlioptas矩阵)。
保证eps嵌入的最小组件数量由下式给出:
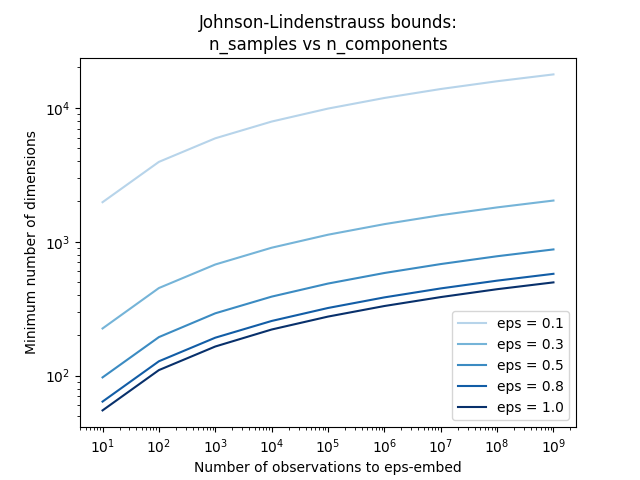
第一个图显示,随着样本数量的增加 n_samples ,最少的维度数量 n_components 为保证 eps - 嵌入。
# range of admissible distortions
eps_range = np.linspace(0.1, 0.99, 5)
colors = plt.cm.Blues(np.linspace(0.3, 1.0, len(eps_range)))
# range of number of samples (observation) to embed
n_samples_range = np.logspace(1, 9, 9)
plt.figure()
for eps, color in zip(eps_range, colors):
min_n_components = johnson_lindenstrauss_min_dim(n_samples_range, eps=eps)
plt.loglog(n_samples_range, min_n_components, color=color)
plt.legend([f"eps = {eps:0.1f}" for eps in eps_range], loc="lower right")
plt.xlabel("Number of observations to eps-embed")
plt.ylabel("Minimum number of dimensions")
plt.title("Johnson-Lindenstrauss bounds:\nn_samples vs n_components")
plt.show()
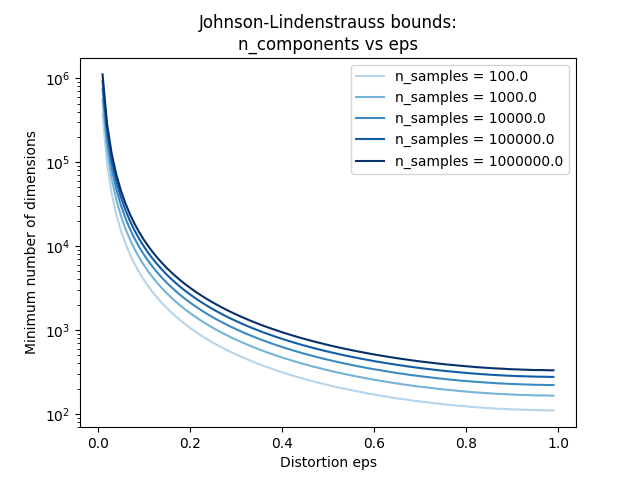
第二个图表明,允许失真的增加 eps 允许大幅减少最少的维度数量 n_components 对于给定数量的样本 n_samples
# range of admissible distortions
eps_range = np.linspace(0.01, 0.99, 100)
# range of number of samples (observation) to embed
n_samples_range = np.logspace(2, 6, 5)
colors = plt.cm.Blues(np.linspace(0.3, 1.0, len(n_samples_range)))
plt.figure()
for n_samples, color in zip(n_samples_range, colors):
min_n_components = johnson_lindenstrauss_min_dim(n_samples, eps=eps_range)
plt.semilogy(eps_range, min_n_components, color=color)
plt.legend([f"n_samples = {n}" for n in n_samples_range], loc="upper right")
plt.xlabel("Distortion eps")
plt.ylabel("Minimum number of dimensions")
plt.title("Johnson-Lindenstrauss bounds:\nn_components vs eps")
plt.show()
经验验证#
我们在20个新闻组文本文档(TF-IDF词频)数据集或数字数据集上验证上述界限:
对于20个新闻组数据集,使用稀疏随机矩阵将总共10万个要素的约300个文档投影到较小的欧几里得空间,该空间具有目标维度数的不同值
n_components.对于数字数据集,300个手写数字图片的一些8 x 8灰度像素数据被随机投影到各种较大维度的空间中
n_components.
默认数据集是20个新闻组数据集。要在数字数据集上运行该示例,请传递 --use-digits-dataset 此脚本的命令行参数。
if "--use-digits-dataset" in sys.argv:
data = load_digits().data[:300]
else:
data = fetch_20newsgroups_vectorized().data[:300]
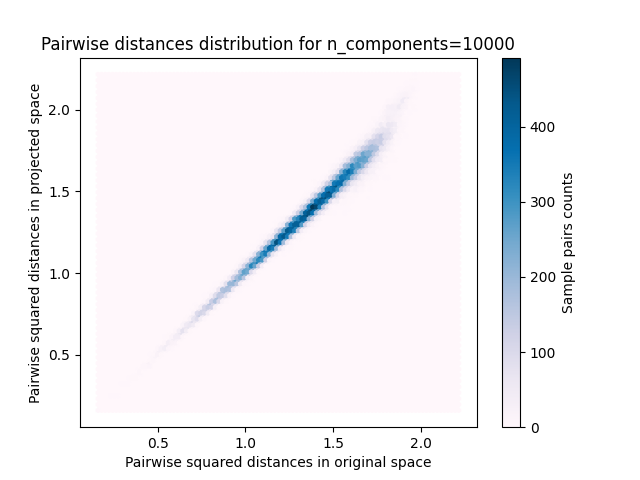
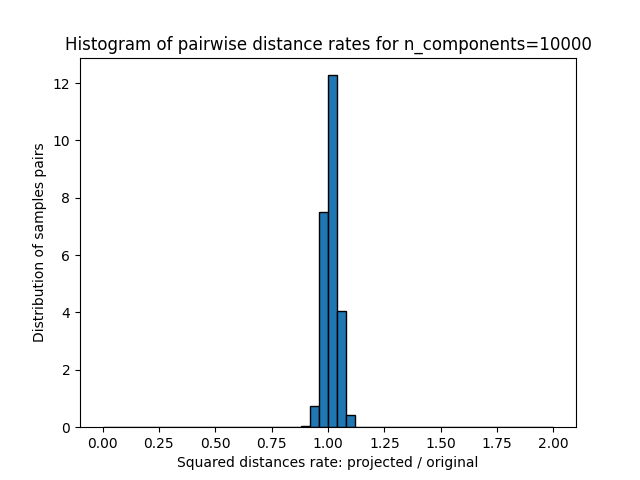
的每个值 n_components ,我们绘制:
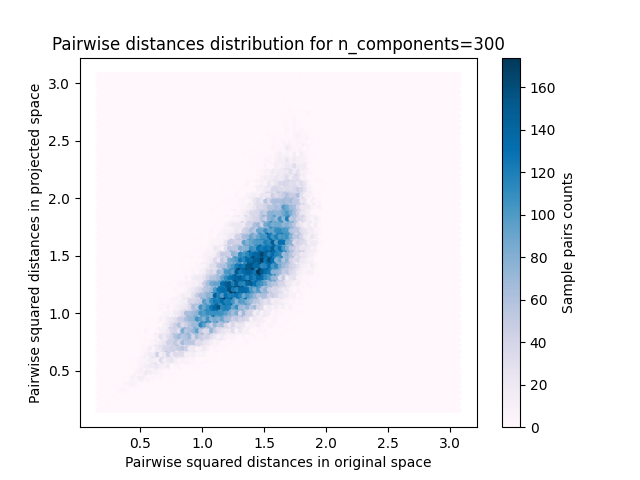
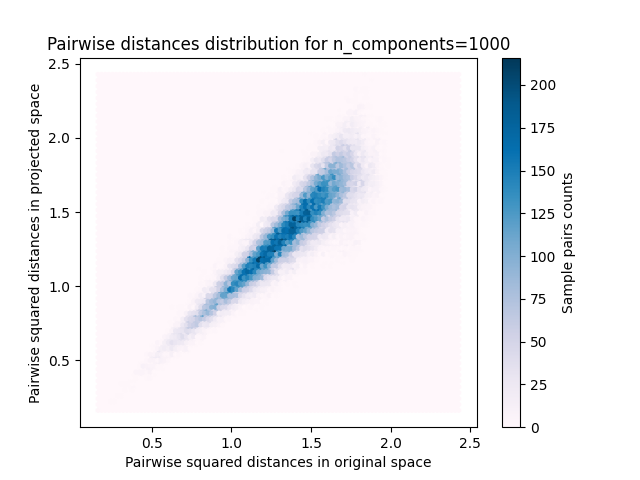
样本对的2D分布,原始空间和投影空间中的成对距离分别为x轴和y轴。
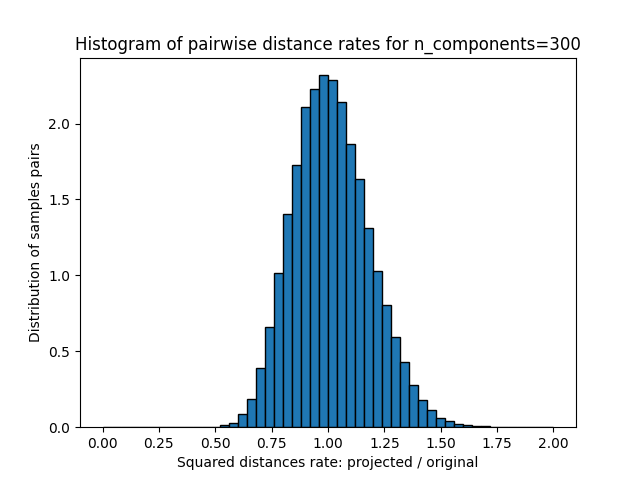
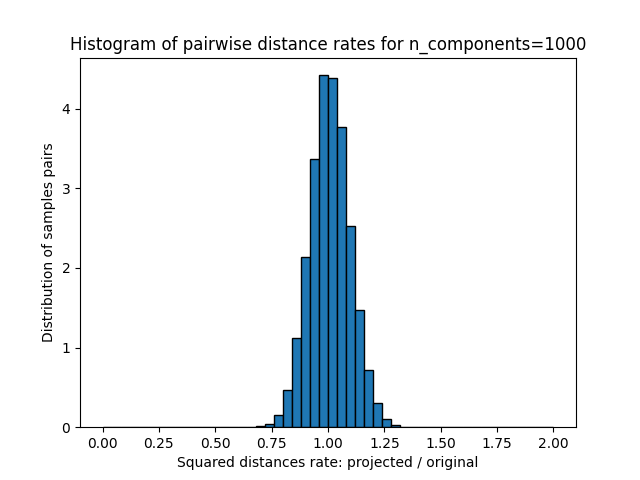
这些距离之比的1D图表(投影/原始)。
n_samples, n_features = data.shape
print(
f"Embedding {n_samples} samples with dim {n_features} using various "
"random projections"
)
n_components_range = np.array([300, 1_000, 10_000])
dists = euclidean_distances(data, squared=True).ravel()
# select only non-identical samples pairs
nonzero = dists != 0
dists = dists[nonzero]
for n_components in n_components_range:
t0 = time()
rp = SparseRandomProjection(n_components=n_components)
projected_data = rp.fit_transform(data)
print(
f"Projected {n_samples} samples from {n_features} to {n_components} in "
f"{time() - t0:0.3f}s"
)
if hasattr(rp, "components_"):
n_bytes = rp.components_.data.nbytes
n_bytes += rp.components_.indices.nbytes
print(f"Random matrix with size: {n_bytes / 1e6:0.3f} MB")
projected_dists = euclidean_distances(projected_data, squared=True).ravel()[nonzero]
plt.figure()
min_dist = min(projected_dists.min(), dists.min())
max_dist = max(projected_dists.max(), dists.max())
plt.hexbin(
dists,
projected_dists,
gridsize=100,
cmap=plt.cm.PuBu,
extent=[min_dist, max_dist, min_dist, max_dist],
)
plt.xlabel("Pairwise squared distances in original space")
plt.ylabel("Pairwise squared distances in projected space")
plt.title("Pairwise distances distribution for n_components=%d" % n_components)
cb = plt.colorbar()
cb.set_label("Sample pairs counts")
rates = projected_dists / dists
print(f"Mean distances rate: {np.mean(rates):.2f} ({np.std(rates):.2f})")
plt.figure()
plt.hist(rates, bins=50, range=(0.0, 2.0), edgecolor="k", density=True)
plt.xlabel("Squared distances rate: projected / original")
plt.ylabel("Distribution of samples pairs")
plt.title("Histogram of pairwise distance rates for n_components=%d" % n_components)
# TODO: compute the expected value of eps and add them to the previous plot
# as vertical lines / region
plt.show()
Embedding 300 samples with dim 130107 using various random projections
Projected 300 samples from 130107 to 300 in 0.310s
Random matrix with size: 1.301 MB
Mean distances rate: 1.02 (0.17)
Projected 300 samples from 130107 to 1000 in 0.882s
Random matrix with size: 4.324 MB
Mean distances rate: 1.01 (0.09)
Projected 300 samples from 130107 to 10000 in 7.199s
Random matrix with size: 43.239 MB
Mean distances rate: 1.01 (0.03)
我们可以看到,对于低价值的 n_components 分布很宽,有许多失真对和倾斜分布(由于左侧零比的硬限制,因为距离总是正值),而对于较大的值 n_components 通过随机投影,失真得到控制,距离得到很好的保留。
言论#
根据JL引理,投影300个样本而没有太多失真将至少需要数千个维度,而无论原始数据集的特征数量如何。
因此,在输入空间中仅具有64个特征的数字数据集上使用随机投影是没有意义的:在这种情况下,它不允许降维。
另一方面,在二十个新闻组中,维度可以从56,436减少到10,000,同时合理地保留成对距离。
Total running time of the script: (0分10.491秒)
相关实例
Gallery generated by Sphinx-Gallery <https://sphinx-gallery.github.io> _