备注
Go to the end 下载完整的示例代码。或者通过浏览器中的MysterLite或Binder运行此示例
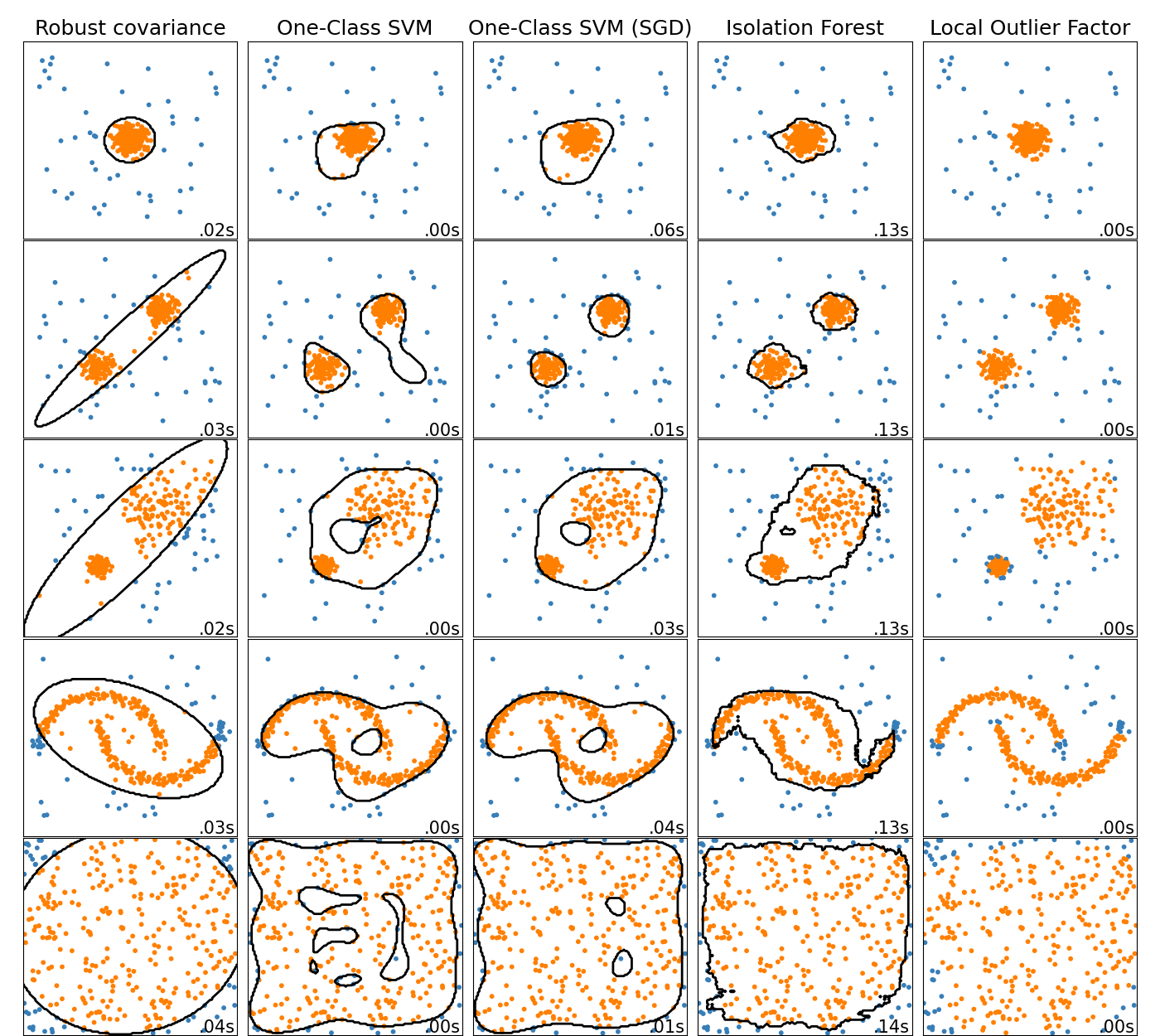
比较玩具数据集异常值检测的异常检测算法#
此示例展示了2D数据集上不同异常检测算法的特征。数据集包含一个或两个模式(高密度区域),以说明算法处理多模式数据的能力。
对于每个数据集,15%的样本作为随机均匀噪音生成。该比例是OneClassSV的nu参数和其他异常值检测算法的污染参数的值。内部值和异常值之间的决策边界以黑色显示,局部异常值因子(LOF)除外,因为当用于异常值检测时,它没有可应用于新数据的预测方法。
的 OneClassSVM 已知对异常值敏感,因此对于异常值检测表现不佳。当训练集不受异常值污染时,该估计器最适合新颖性检测。也就是说,在多维中或在不对内部数据的分布进行任何假设的情况下进行异常值检测是非常具有挑战性的,并且一级支持机可能会在这些情况下给出有用的结果,具体取决于其超参数的值。
的 sklearn.linear_model.SGDOneClassSVM 是基于随机梯度下降(BCD)的一类支持者支持者的实现。与核逼近相结合,该估计器可以用于逼近核化的解 sklearn.svm.OneClassSVM .我们注意到,尽管不同,但 sklearn.linear_model.SGDOneClassSVM 以及那些 sklearn.svm.OneClassSVM 非常相似。使用的主要优势 sklearn.linear_model.SGDOneClassSVM 它与样本数量呈线性缩放。
sklearn.covariance.EllipticEnvelope 假设数据是高斯数据并学习椭圆。因此,当数据不是单峰时,它会降级。然而请注意,该估计器对离群值具有鲁棒性。
IsolationForest and
LocalOutlierFactor seem to perform reasonably well
for multi-modal data sets. The advantage of
LocalOutlierFactor over the other estimators is
shown for the third data set, where the two modes have different densities.
This advantage is explained by the local aspect of LOF, meaning that it only
compares the score of abnormality of one sample with the scores of its
neighbors.
最后,对于最后一个数据集,很难说一个样本比另一个样本更异常,因为它们均匀分布在超立方体中。除了 OneClassSVM 这有点过于适合,所有估计器都为这种情况提供了不错的解决方案。在这种情况下,明智的做法是更仔细地观察样本的异常评分,因为好的估计者应该为所有样本分配相似的评分。
虽然这些例子提供了一些关于算法的直觉,但这种直觉可能不适用于非常高维度的数据。
最后,请注意,模型的参数是这里精心挑选的,但在实践中需要调整。在缺乏标记数据的情况下,该问题是完全无监督的,因此模型选择可能是一个挑战。
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
import time
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
from sklearn import svm
from sklearn.covariance import EllipticEnvelope
from sklearn.datasets import make_blobs, make_moons
from sklearn.ensemble import IsolationForest
from sklearn.kernel_approximation import Nystroem
from sklearn.linear_model import SGDOneClassSVM
from sklearn.neighbors import LocalOutlierFactor
from sklearn.pipeline import make_pipeline
matplotlib.rcParams["contour.negative_linestyle"] = "solid"
# Example settings
n_samples = 300
outliers_fraction = 0.15
n_outliers = int(outliers_fraction * n_samples)
n_inliers = n_samples - n_outliers
# define outlier/anomaly detection methods to be compared.
# the SGDOneClassSVM must be used in a pipeline with a kernel approximation
# to give similar results to the OneClassSVM
anomaly_algorithms = [
(
"Robust covariance",
EllipticEnvelope(contamination=outliers_fraction, random_state=42),
),
("One-Class SVM", svm.OneClassSVM(nu=outliers_fraction, kernel="rbf", gamma=0.1)),
(
"One-Class SVM (SGD)",
make_pipeline(
Nystroem(gamma=0.1, random_state=42, n_components=150),
SGDOneClassSVM(
nu=outliers_fraction,
shuffle=True,
fit_intercept=True,
random_state=42,
tol=1e-6,
),
),
),
(
"Isolation Forest",
IsolationForest(contamination=outliers_fraction, random_state=42),
),
(
"Local Outlier Factor",
LocalOutlierFactor(n_neighbors=35, contamination=outliers_fraction),
),
]
# Define datasets
blobs_params = dict(random_state=0, n_samples=n_inliers, n_features=2)
datasets = [
make_blobs(centers=[[0, 0], [0, 0]], cluster_std=0.5, **blobs_params)[0],
make_blobs(centers=[[2, 2], [-2, -2]], cluster_std=[0.5, 0.5], **blobs_params)[0],
make_blobs(centers=[[2, 2], [-2, -2]], cluster_std=[1.5, 0.3], **blobs_params)[0],
4.0
* (
make_moons(n_samples=n_samples, noise=0.05, random_state=0)[0]
- np.array([0.5, 0.25])
),
14.0 * (np.random.RandomState(42).rand(n_samples, 2) - 0.5),
]
# Compare given classifiers under given settings
xx, yy = np.meshgrid(np.linspace(-7, 7, 150), np.linspace(-7, 7, 150))
plt.figure(figsize=(len(anomaly_algorithms) * 2 + 4, 12.5))
plt.subplots_adjust(
left=0.02, right=0.98, bottom=0.001, top=0.96, wspace=0.05, hspace=0.01
)
plot_num = 1
rng = np.random.RandomState(42)
for i_dataset, X in enumerate(datasets):
# Add outliers
X = np.concatenate([X, rng.uniform(low=-6, high=6, size=(n_outliers, 2))], axis=0)
for name, algorithm in anomaly_algorithms:
t0 = time.time()
algorithm.fit(X)
t1 = time.time()
plt.subplot(len(datasets), len(anomaly_algorithms), plot_num)
if i_dataset == 0:
plt.title(name, size=18)
# fit the data and tag outliers
if name == "Local Outlier Factor":
y_pred = algorithm.fit_predict(X)
else:
y_pred = algorithm.fit(X).predict(X)
# plot the levels lines and the points
if name != "Local Outlier Factor": # LOF does not implement predict
Z = algorithm.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z, levels=[0], linewidths=2, colors="black")
colors = np.array(["#377eb8", "#ff7f00"])
plt.scatter(X[:, 0], X[:, 1], s=10, color=colors[(y_pred + 1) // 2])
plt.xlim(-7, 7)
plt.ylim(-7, 7)
plt.xticks(())
plt.yticks(())
plt.text(
0.99,
0.01,
("%.2fs" % (t1 - t0)).lstrip("0"),
transform=plt.gca().transAxes,
size=15,
horizontalalignment="right",
)
plot_num += 1
plt.show()
Total running time of the script: (0分3.219秒)
相关实例
Gallery generated by Sphinx-Gallery <https://sphinx-gallery.github.io> _